import numpy as np
import scipy
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
import sys
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
sns.set_context('notebook', font_scale=1.5)
sns.set_style('whitegrid')
%config InlineBackend.figure_format = 'retina'
import tensorflow as tf
print(f'python version {sys.version[:5]}')
print(f'Tensorflow version {tf.__version__}')
print(f'Eager execution on: {tf.executing_eagerly()}')
import tensorflow_probability as tfp
from tensorflow_probability import distributions as tfd
if tf.test.gpu_device_name() != '/device:GPU:0':
print('WARNING: GPU device not found.')
else:
print(f'SUCCESS: Found GPU: {tf.test.gpu_device_name()}')
python version 3.7.6
Tensorflow version 2.2.0
Eager execution on: True
WARNING: GPU device not found.
Generate data
\[
\begin{align}
y & \sim N(\mu, \sigma) \\\\
\mu & = f(x_1, x_2) \\\\
f(x_1, x_2) & = \sqrt{x_1^2 + x_2^2} \\\\
x_1 & = -4.5 \\\\
x_2 & = 5.5 \\\\
\sigma & = 2
\end{align}
\]
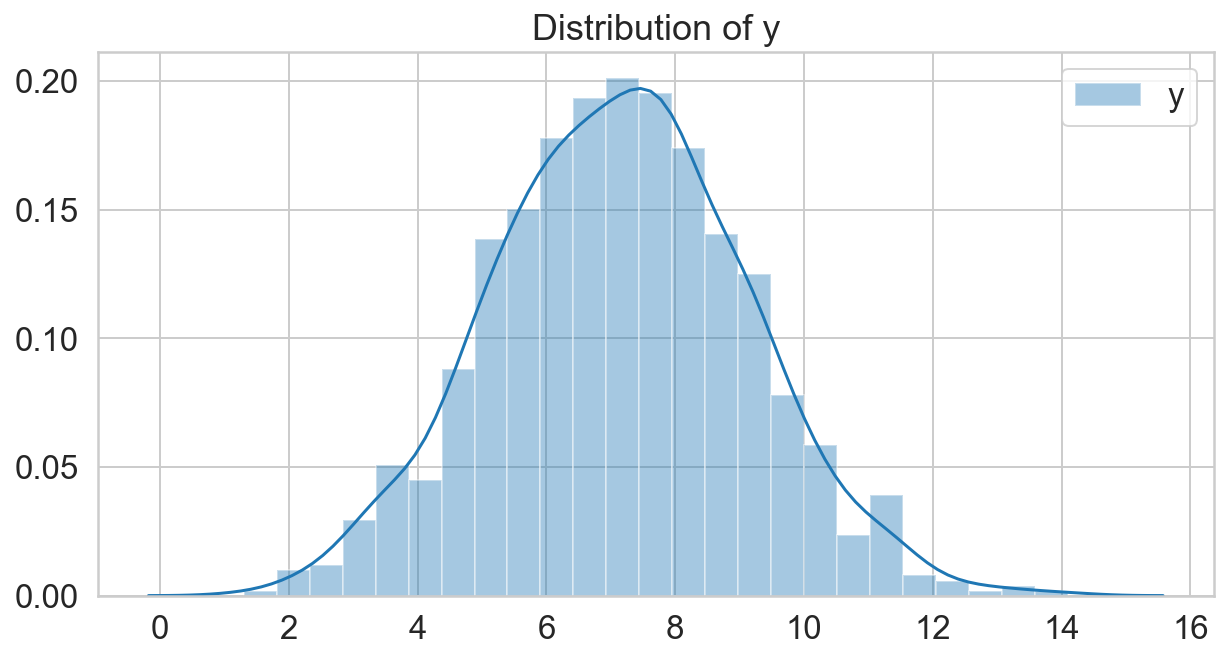
np.random.seed(30)
N = 1000
x_1 = -4.5
x_2 = 5.5
mu = np.sqrt(x_1**2 + x_2**2)
sigma = 2
y = scipy.stats.norm(mu, sigma).rvs(N)
fig, ax = plt.subplots(figsize=(10, 5));
sns.distplot(y, label='y');
ax.legend();
ax.set_title('Distribution of y');
Statistical Model: prior and likelihood function
\[
\begin{align}
y & \sim N(\mu, \sigma) \\\\
\mu & = f(x_1, x_2) \\\\
f(x_1, x_2) & = \sqrt{x_1^2 + x_2^2} \\\\
x_1 & \sim Uniform(-10, 10) \\\\
x_2 & \sim Uniform(-10, 10) \\\\
\sigma & \sim Exponential(1)
\end{align}
\]
def unnormalized_log_prob(theta):
# theta = [sigma, x1, x2]
sigma = theta[0]
mu = tf.norm(theta[1:])
loglikelihood = tf.reduce_sum(tfd.Normal(mu, sigma).log_prob(y))
logprior = tfd.Exponential(1).log_prob(sigma)
logprior += tf.reduce_sum(tfd.Uniform(-10, 10).log_prob(theta[1:]))
logposterior = logprior + loglikelihood
return logposterior
theta = tf.constant([1, 1, 1], dtype=tf.float32)
logposterior = unnormalized_log_prob(theta)
## Run HMC
starting_state = tf.constant([1, 1, 1], dtype=tf.float32)
num_results = int(1e5)
num_burnin_steps = int(1e3)
adaptive_hmc = tfp.mcmc.SimpleStepSizeAdaptation(
tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_log_prob,
num_leapfrog_steps=3,
step_size=1.),
num_adaptation_steps=int(num_burnin_steps * 0.8))
# Run the chain (with burn-in).
@tf.function
def run_chain():
samples, is_accepted = tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=starting_state,
kernel=adaptive_hmc,
trace_fn=lambda _, pkr: pkr.inner_results.is_accepted)
return samples, is_accepted
samples, is_accepted = run_chain()
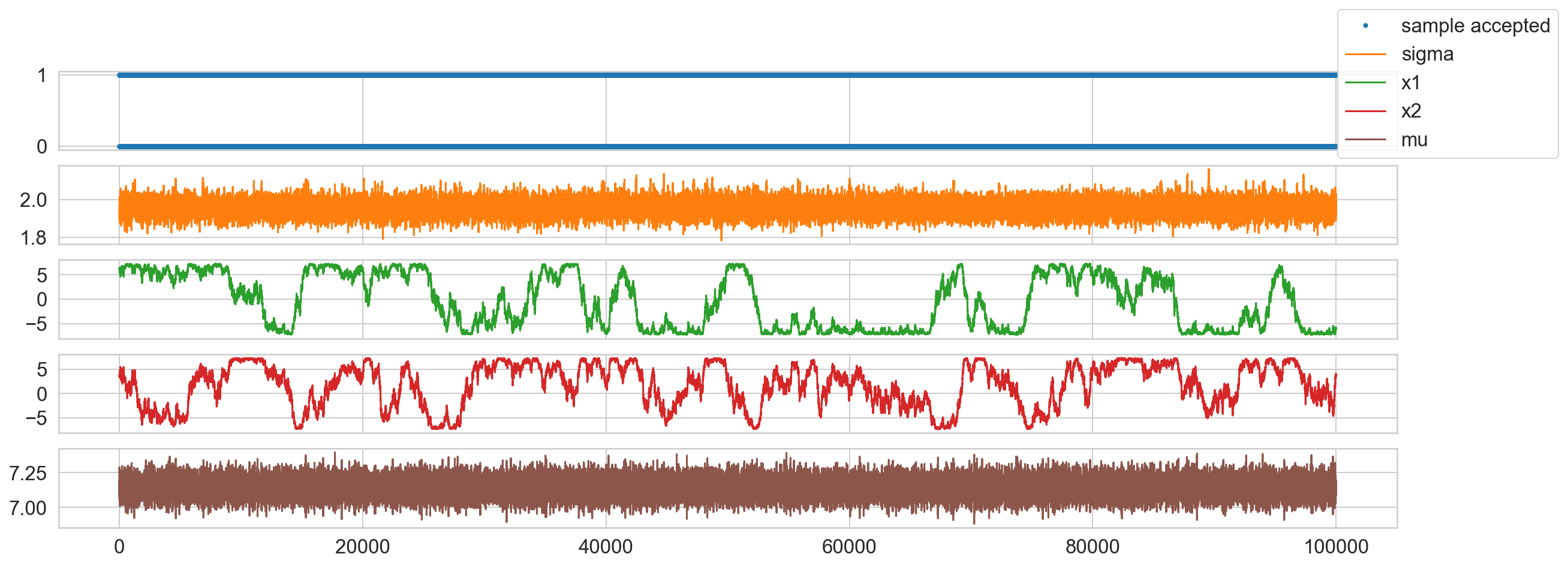
# Plot Samples
accepted_samples = samples[is_accepted.numpy()]
print(f'Accepted samples: {len(accepted_samples):,}/{len(samples):,}')
fig, ax = plt.subplots(5, 1, sharex=True, figsize=(20, 7));
ax[0].plot(is_accepted.numpy(), '.', c=sns.color_palette()[0], label='sample accepted');
ax[1].plot(samples[:, 0], c=sns.color_palette()[1], label='sigma');
ax[2].plot(samples[:, 1], c=sns.color_palette()[2], label='x1');
ax[3].plot(samples[:, 2], c=sns.color_palette()[3], label='x2');
ax[4].plot(tf.sqrt(tf.reduce_sum(samples[:, 1:]**2, axis=1)), c=sns.color_palette()[5], label='mu');
fig.legend();
Accepted samples: 71,781/100,000
s_sigma = accepted_samples[:, 0]
s_x1 = accepted_samples[:, 1]
s_x2 = accepted_samples[:, 2]
s_mu = np.sqrt(s_x1**2 + s_x2**2)
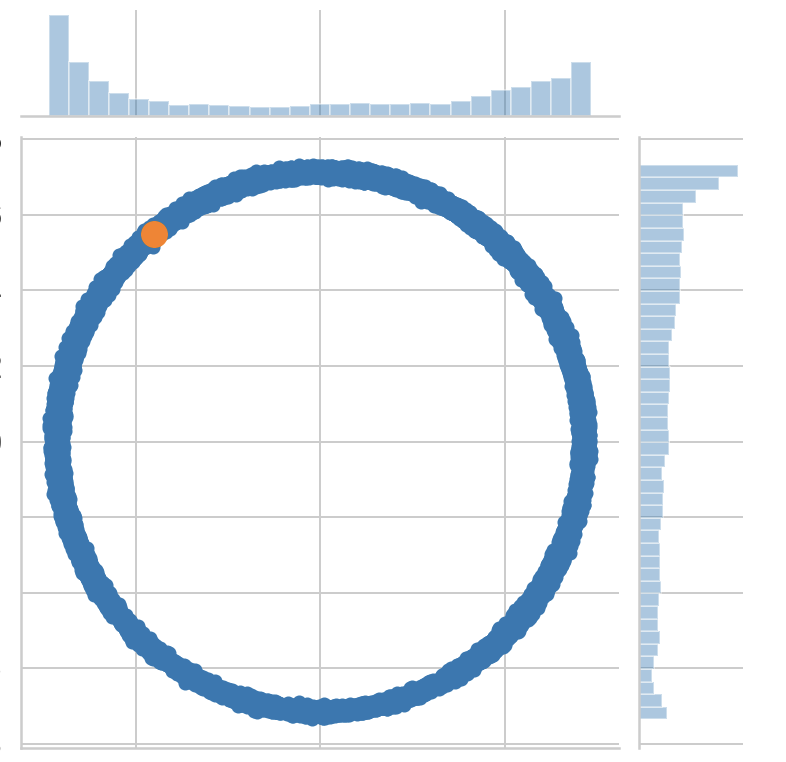
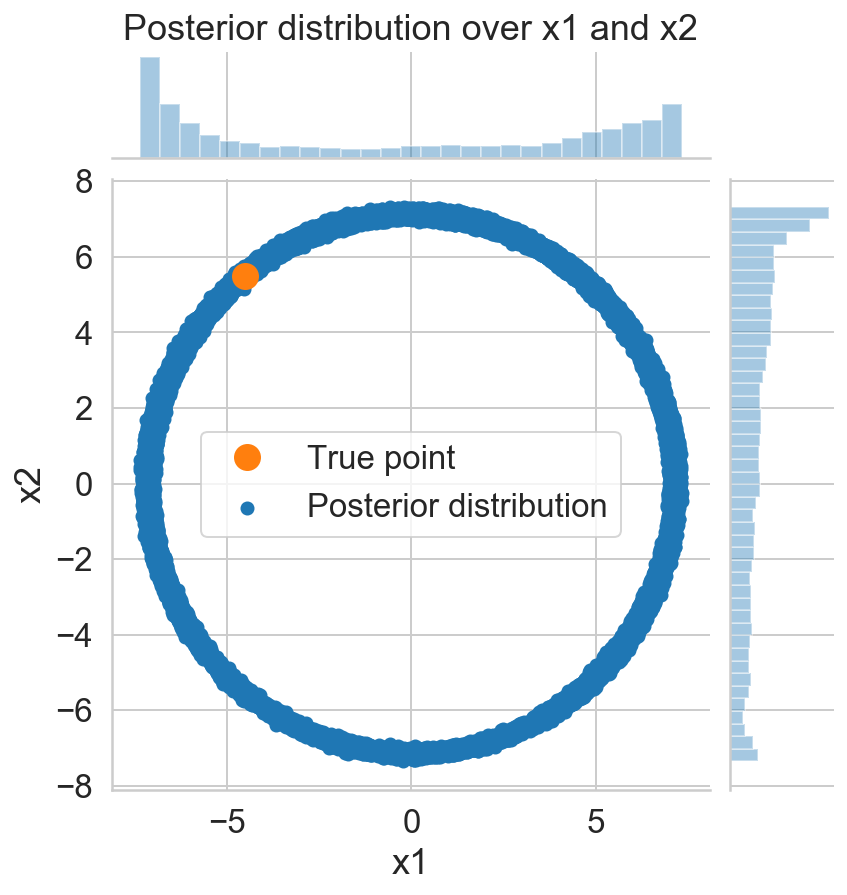
p = sns.jointplot(s_x1, s_x2, label='Posterior distribution');
ax = p.fig.axes[0];
ax.set_xlabel('x1')
ax.set_ylabel('x2')
ax.plot(x_1, x_2, '.', markersize=25, c=sns.color_palette()[1], label='True point');
ax.legend();
p.fig.axes[1].set_title('Posterior distribution over x1 and x2');
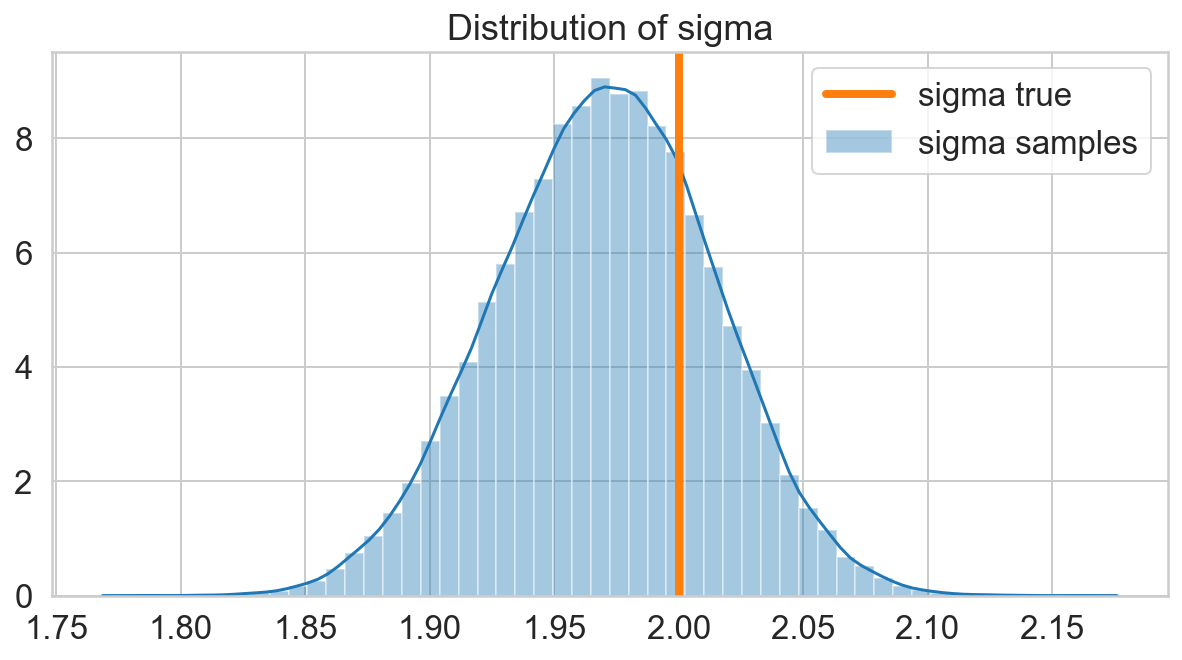
fig, ax = plt.subplots(figsize=(10, 5))
sns.distplot(s_sigma, ax=ax, label='sigma samples')
ax.axvline(x=sigma, lw=4, c=sns.color_palette()[1], label='sigma true')
ax.set_title('Distribution of sigma');
ax.legend();
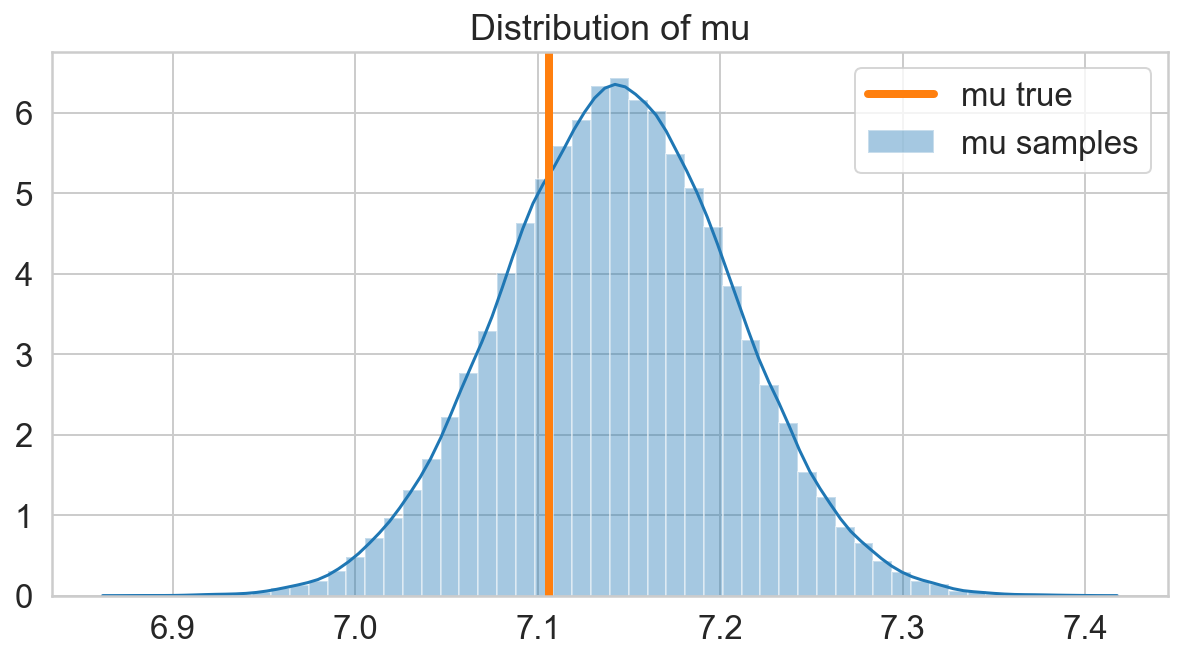
fig, ax = plt.subplots(figsize=(10, 5));
sns.distplot(s_mu, label='mu samples', ax=ax);
ax.axvline(x=mu, lw=4, c=sns.color_palette()[1], label='mu true')
ax.legend();
ax.set_title('Distribution of mu');
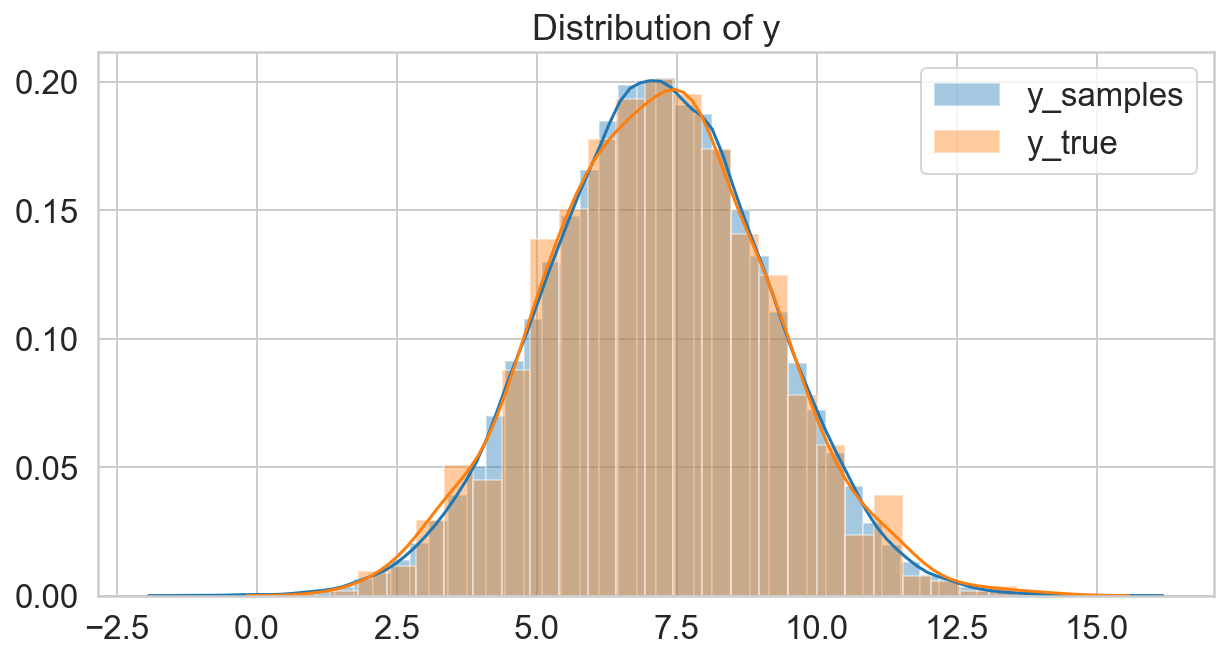
y_samples = scipy.stats.norm(s_mu, s_sigma).rvs()
fig, ax = plt.subplots(figsize=(10, 5));
sns.distplot(y_samples, label='y_samples', ax=ax);
sns.distplot(y, label='y_true', ax=ax);
ax.legend();
ax.set_title('Distribution of y');